3. Algorithms¶
The algorithms module is intended to contain some specific algorithms in order to execute very common evolutionary algorithms. The method used here are more for convenience than reference as the implementation of every evolutionary algorithm may vary infinitely. Most of the algorithms in this module use operators registered in the toolbox. Generaly, the keyword used are mate() for crossover, mutate() for mutation, select() for selection and evaluate() for evaluation.
You are encouraged to write your own algorithms in order to make them do what you really want them to do.
- deap.algorithms.eaSimple(toolbox, population, cxpb, mutpb, ngen[, stats, halloffame])¶
This algorithm reproduce the simplest evolutionary algorithm as presented in chapter 7 of Back, Fogel and Michalewicz, “Evolutionary Computation 1 : Basic Algorithms and Operators”, 2000. It uses
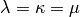 and goes as follow.
It first initializes the population (
and goes as follow.
It first initializes the population (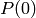 ) by evaluating
every individual presenting an invalid fitness. Then, it enters the
evolution loop that begins by the selection of the
) by evaluating
every individual presenting an invalid fitness. Then, it enters the
evolution loop that begins by the selection of the 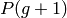 population. Then the crossover operator is applied on a proportion of
population. Then the crossover operator is applied on a proportion of
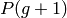 according to the cxpb probability, the resulting and the
untouched individuals are placed in
according to the cxpb probability, the resulting and the
untouched individuals are placed in 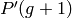 . Thereafter, a
proportion of
. Thereafter, a
proportion of 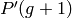 , determined by mutpb, is
mutated and placed in
, determined by mutpb, is
mutated and placed in 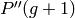 , the untouched individuals are
transferred
, the untouched individuals are
transferred 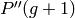 . Finally, those new individuals are evaluated
and the evolution loop continues until ngen generations are completed.
Briefly, the operators are applied in the following order
. Finally, those new individuals are evaluated
and the evolution loop continues until ngen generations are completed.
Briefly, the operators are applied in the following orderevaluate(population) for i in range(ngen): offspring = select(population) offspring = mate(offspring) offspring = mutate(offspring) evaluate(offspring) population = offspring
This function expects toolbox.mate(), toolbox.mutate(), toolbox.select() and toolbox.evaluate() aliases to be registered in the toolbox.
- deap.algorithms.varSimple(toolbox, population, cxpb, mutpb)¶
Part of the eaSimple() algorithm applying only the variation part (crossover followed by mutation). The modified individuals have their fitness invalidated. The individuals are not cloned so there can be twice a reference to the same individual.
This function expects toolbox.mate() and toolbox.mutate() aliases to be registered in the toolbox.
- deap.algorithms.varAnd(toolbox, population, cxpb, mutpb)¶
Part of an evolutionary algorithm applying only the variation part (crossover and mutation). The modified individuals have their fitness invalidated. The individuals are cloned so returned population is independent of the input population.
The variator goes as follow. First, the parental population
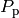 is duplicated using the toolbox.clone() method
and the result is put into the offspring population
is duplicated using the toolbox.clone() method
and the result is put into the offspring population 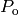 .
A first loop over
.
A first loop over 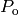 is executed to mate consecutive
individuals. According to the crossover probability cxpb, the
individuals
is executed to mate consecutive
individuals. According to the crossover probability cxpb, the
individuals 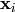 and
and 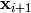 are mated
using the toolbox.mate() method. The resulting children
are mated
using the toolbox.mate() method. The resulting children
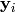 and
and 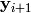 replace their respective
parents in
replace their respective
parents in 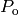 . A second loop over the resulting
. A second loop over the resulting
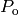 is executed to mutate every individual with a
probability mutpb. When an individual is mutated it replaces its not
mutated version in
is executed to mutate every individual with a
probability mutpb. When an individual is mutated it replaces its not
mutated version in 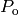 . The resulting
. The resulting
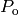 is returned.
is returned.This variation is named And beceause of its propention to apply both crossover and mutation on the individuals. Both probabilities should be in
![[0, 1]](../_images/math/ab178d831a786b92cb4c9ddc2d33578223036f98.png) .
.
- deap.algorithms.eaMuPlusLambda(toolbox, population, mu, lambda_, cxpb, mutpb, ngen[, stats, halloffame])¶
This is the
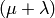 evolutionary algorithm. First,
the individuals having an invalid fitness are evaluated. Then, the
evolutionary loop begins by producing lambda offspring from the
population, the offspring are generated by a crossover, a mutation or a
reproduction proportionally to the probabilities cxpb, mutpb and
1 - (cxpb + mutpb). The offspring are then evaluated and the next
generation population is selected from both the offspring and the
population. Briefly, the operators are applied as following
evolutionary algorithm. First,
the individuals having an invalid fitness are evaluated. Then, the
evolutionary loop begins by producing lambda offspring from the
population, the offspring are generated by a crossover, a mutation or a
reproduction proportionally to the probabilities cxpb, mutpb and
1 - (cxpb + mutpb). The offspring are then evaluated and the next
generation population is selected from both the offspring and the
population. Briefly, the operators are applied as followingevaluate(population) for i in range(ngen): offspring = generate_offspring(population) evaluate(offspring) population = select(population + offspring)
This function expects toolbox.mate(), toolbox.mutate(), toolbox.select() and toolbox.evaluate() aliases to be registered in the toolbox.
Note
Both produced individuals from a crossover are put in the offspring pool.
- deap.algorithms.eaMuCommaLambda(toolbox, population, mu, lambda_, cxpb, mutpb, ngen[, stats, halloffame])¶
This is the
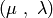 evolutionary algorithm. First,
the individuals having an invalid fitness are evaluated. Then, the
evolutionary loop begins by producing lambda offspring from the
population, the offspring are generated by a crossover, a mutation or a
reproduction proportionally to the probabilities cxpb, mutpb and
1 - (cxpb + mutpb). The offspring are then evaluated and the next
generation population is selected only from the offspring. Briefly,
the operators are applied as following
evolutionary algorithm. First,
the individuals having an invalid fitness are evaluated. Then, the
evolutionary loop begins by producing lambda offspring from the
population, the offspring are generated by a crossover, a mutation or a
reproduction proportionally to the probabilities cxpb, mutpb and
1 - (cxpb + mutpb). The offspring are then evaluated and the next
generation population is selected only from the offspring. Briefly,
the operators are applied as followingevaluate(population) for i in range(ngen): offspring = generate_offspring(population) evaluate(offspring) population = select(offspring)
This function expects toolbox.mate(), toolbox.mutate(), toolbox.select() and toolbox.evaluate() aliases to be registered in the toolbox.
Note
Both produced individuals from the crossover are put in the offspring pool.
- deap.algorithms.varLambda(toolbox, population, lambda_, cxpb, mutpb)¶
Part of the eaMuPlusLambda() and eaMuCommaLambda() algorithms that produce the lambda new individuals. The modified individuals have their fitness invalidated. The individuals are not cloned so there can be twice a reference to the same individual.
This function expects toolbox.mate() and toolbox.mutate() aliases to be registered in the toolbox.
- deap.algorithms.varOr(toolbox, population, lambda_, cxpb, mutpb)¶
Part of an evolutionary algorithm applying only the variation part (crossover, mutation or reproduction). The modified individuals have their fitness invalidated. The individuals are cloned so returned population is independent of the input population.
The variator goes as follow. On each of the lambda_ iteration, it selects one of the three operations; crossover, mutation or reproduction. In the case of a crossover, two individuals are selected at random from the parental population
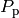 , those individuals are cloned
using the toolbox.clone() method and then mated using the
toolbox.mate() method. Only the first child is appended to the
offspring population
, those individuals are cloned
using the toolbox.clone() method and then mated using the
toolbox.mate() method. Only the first child is appended to the
offspring population 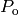 , the second child is discarded.
In the case of a mutation, one individual is selected at random from
, the second child is discarded.
In the case of a mutation, one individual is selected at random from
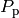 , it is cloned and then mutated using using the
toolbox.mutate() method. The resulting mutant is appended to
, it is cloned and then mutated using using the
toolbox.mutate() method. The resulting mutant is appended to
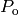 . In the case of a reproduction, one individual is
selected at random from
. In the case of a reproduction, one individual is
selected at random from 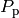 , cloned and appended to
, cloned and appended to
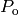 .
.This variation is named Or beceause an offspring will never result from both operations crossover and mutation. The sum of both probabilities shall be in
![[0, 1]](../_images/math/ab178d831a786b92cb4c9ddc2d33578223036f98.png) , the reproduction probability is
1 - cxpb - mutpb.
, the reproduction probability is
1 - cxpb - mutpb.
- deap.algorithms.eaSteadyState(toolbox, population, ngen[, stats, halloffame])¶
The steady-state evolutionary algorithm. Every generation, a single new individual is produced and put in the population producing a population of size
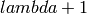 , then
, then 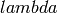 individuals are kept according
to the selection operator present in the toolbox.
individuals are kept according
to the selection operator present in the toolbox.This function expects toolbox.mate(), toolbox.mutate(), toolbox.select() and toolbox.evaluate() aliases to be registered in the toolbox.
- deap.algorithms.varSteadyState(toolbox, population)¶
Part of the eaSteadyState() algorithm that produce the new individual by crossover of two randomly selected parents and mutation on one randomly selected child. The modified individual has its fitness invalidated. The individuals are not cloned so there can be twice a reference to the same individual.
This function expects toolbox.mate(), toolbox.mutate() and toolbox.select() aliases to be registered in the toolbox.
4. Covariance Matrix Adaptation Evolution Strategy¶
A module that provides support for the Covariance Matrix Adaptation Evolution Strategy.
- deap.cma.esCMA(toolbox, population, sigma, ngen[, halloffame, **kargs])¶
The CMA-ES algorithm as described in Hansen, N. (2006). The CMA Evolution Strategy: A Comparing Rewiew.
The provided population should be a list of one or more individuals.
- class deap.cma.CMAStrategy(population, sigma[, params])¶
Additional configuration may be passed through the params argument as a dictionary,
Parameter Default Details lambda_ int(4 + 3 * log(N)) Number of children to produce at each generation, N is the individual’s size (integer). mu int(lambda_ / 2) The number of parents to keep from the lambda children (integer). weights "superlinear" Decrease speed, can be "superlinear", "linear" or "equal". cs (mueff + 2) / (N + mueff + 3) Cumulation constant for step-size. damps 1 + 2 * max(0, sqrt(( mueff - 1) / (N + 1)) - 1) + cs Damping for step-size. ccum 4 / (N + 4) Cumulation constant for covariance matrix. ccov1 2 / ((N + 1.3)^2 + mueff) Learning rate for rank-one update. ccovmu 2 * (mueff - 2 + 1 / mueff) / ((N + 2)^2 + mueff) Learning rate for rank-mu update. - computeParams(params)¶
Those parameters depends on lambda and need to computed again if it changes during evolution.
- generate(ind_init)¶
Generate a population from the current strategy using the centroid individual as parent.
- update(population)¶
Update the current covariance matrix strategy.