The tools module contains the operators for evolutionary algorithms. They are used to modify, select and move the individuals in their environment. The set of operators it contains are readily usable in the Toolbox. In addition to the basic operators this module also contains utility tools to enhance the basic algorithms with Statistics, HallOfFame, Checkpoint, and History.
The operator set does the minimum job for transforming or selecting individuals. This means, for example, that providing two individuals to the crossover will transform those individuals in-place. The responsibility of making offspring(s) independent of their parent(s) and invalidating the fitness is left to the user and is generally fulfilled in the algorithms by calling toolbox.clone() on an individual to duplicate it and del on the values attribute of the individual’s fitness to invalidate it.
Here is a list of the implemented operators in DEAP,
and genetic programming specific operators.
| Initialization | Crossover | Mutation | Bloat control |
|---|---|---|---|
| genFull() | cxOnePoint() | mutShrink() | staticLimit() |
| genGrow() | cxOnePointLeafBiased() | mutUniform() | selDoubleTournament() |
| genHalfAndHalf() | mutNodeReplacement() | ||
| mutEphemeral() | |||
| mutInsert() |
Call the function container with a generator function corresponding to the calling n times the function func.
| Parameters: |
|
|---|---|
| Returns: | An instance of the container filled with data from func. |
This helper function can can be used in conjunction with a Toolbox to register a generator of filled containers, as individuals or population.
>>> initRepeat(list, random.random, 2)
...
[0.4761..., 0.6302...]
See the List of Floats and Population tutorials for more examples.
Call the function container with an iterable as its only argument. The iterable must be returned by the method or the object generator.
| Parameters: |
|
|---|---|
| Returns: | An instance of the container filled with data from the generator. |
This helper function can can be used in conjunction with a Toolbox to register a generator of filled containers, as individuals or population.
>>> from random import sample
>>> from functools import partial
>>> gen_idx = partial(sample, range(10), 10)
>>> initIterate(list, gen_idx)
[4, 5, 3, 6, 0, 9, 2, 7, 1, 8]
See the Permutation and Arithmetic Expression tutorials for more examples.
Call the function container with a generator function corresponding to the calling n times the functions present in seq_func.
| Parameters: |
|
|---|---|
| Returns: | An instance of the container filled with data from the returned by the functions. |
This helper function can can be used in conjunction with a Toolbox to register a generator of filled containers, as individuals or population.
>>> func_seq = [lambda:1 , lambda:'a', lambda:3]
>>> initCycle(list, func_seq, n=2)
[1, 'a', 3, 1, 'a', 3]
See the A Funky One tutorial for an example.
Generate an expression where each leaf has a the same depth between min and max.
| Parameters: |
|
|---|---|
| Returns: | A full tree with all leaves at the same depth. |
Generate an expression where each leaf might have a different depth between min and max.
| Parameters: |
|
|---|---|
| Returns: | A grown tree with leaves at possibly different depths. |
Generate an expression with a PrimitiveSet pset. Half the time, the expression is generated with genGrow(), the other half, the expression is generated with genFull().
| Parameters: |
|
|---|---|
| Returns: | Either, a full or a grown tree. |
Deprecated since version 1.0: The function has been renamed. Use genHalfAndHalf() instead.
Executes a one point crossover on the input sequence individuals. The two individuals are modified in place. The resulting individuals will respectively have the length of the other.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the randint() function from the python base random module.
Executes a two-point crossover on the input sequence individuals. The two individuals are modified in place and both keep their original length.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the randint() function from the Python base random module.
Deprecated since version 1.0: The function has been renamed. Use cxTwoPoint() instead.
Executes a uniform crossover that modify in place the two sequence individuals. The attributes are swapped accordingto the indpb probability.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the random() function from the python base random module.
Executes a partially matched crossover (PMX) on the input individuals. The two individuals are modified in place. This crossover expects sequence individuals of indices, the result for any other type of individuals is unpredictable.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
Moreover, this crossover generates two children by matching pairs of values in a certain range of the two parents and swapping the values of those indexes. For more details see [Goldberg1985].
This function uses the randint() function from the python base random module.
| [Goldberg1985] | Goldberg and Lingel, “Alleles, loci, and the traveling salesman problem”, 1985. |
Executes a uniform partially matched crossover (UPMX) on the input individuals. The two individuals are modified in place. This crossover expects sequence individuals of indices, the result for any other type of individuals is unpredictable.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
Moreover, this crossover generates two children by matching pairs of values chosen at random with a probability of indpb in the two parents and swapping the values of those indexes. For more details see [Cicirello2000].
This function uses the random() and randint() functions from the python base random module.
| [Cicirello2000] | Cicirello and Smith, “Modeling GA performance for control parameter optimization”, 2000. |
Executes an ordered crossover (OX) on the input individuals. The two individuals are modified in place. This crossover expects sequence individuals of indices, the result for any other type of individuals is unpredictable.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
Moreover, this crossover generates holes in the input individuals. A hole is created when an attribute of an individual is between the two crossover points of the other individual. Then it rotates the element so that all holes are between the crossover points and fills them with the removed elements in order. For more details see [Goldberg1989].
This function uses the sample() function from the python base random module.
| [Goldberg1989] | Goldberg. Genetic algorithms in search, optimization and machine learning. Addison Wesley, 1989 |
Executes a blend crossover that modify in-place the input individuals. The blend crossover expects sequence individuals of floating point numbers.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the random() function from the python base random module.
Executes a blend crossover on both, the individual and the strategy. The individuals shall be a sequence and must have a sequence strategy attribute. Adjustement of the minimal strategy shall be done after the call to this function, consider using a decorator.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two evolution strategies. |
This function uses the random() function from the python base random module.
Executes a classical two points crossover on both the individuals and their strategy. The individuals shall be a sequence and must have a sequence strategy attribute. The crossover points for the individual and the strategy are the same.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two evolution strategies. |
This function uses the randint() function from the python base random module.
Deprecated since version 1.0: The function has been renamed. Use cxESTwoPoint() instead.
Executes a simulated binary crossover that modify in-place the input individuals. The simulated binary crossover expects sequence individuals of floating point numbers.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the random() function from the python base random module.
Executes a simulated binary crossover that modify in-place the input individuals. The simulated binary crossover expects sequence individuals of floating point numbers.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the random() function from the python base random module.
Note
This implementation is similar to the one implemented in the original NSGA-II C code presented by Deb.
Executes a one point crossover on sequence individual. The crossover will in most cases change the individuals size. The two individuals are modified in place.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two individuals. |
This function uses the randint() function from the python base random module.
Randomly select in each individual and exchange each subtree with the point as root between each individual.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two trees. |
Randomly select crossover point in each individual and exchange each subtree with the point as root between each individual.
| Parameters: |
|
|---|---|
| Returns: | A tuple of two typed trees. |
When the nodes are strongly typed, the operator makes sure the second node type corresponds to the first node type.
The parameter termpb sets the probability to choose between a terminal or non-terminal crossover point. For instance, as defined by Koza, non- terminal primitives are selected for 90% of the crossover points, and terminals for 10%, so termpb should be set to 0.1.
This function applies a gaussian mutation of mean mu and standard deviation sigma on the input individual. This mutation expects a sequence individual composed of real valued attributes. The indpb argument is the probability of each attribute to be mutated.
| Parameters: | |
|---|---|
| Returns: | A tuple of one individual. |
This function uses the random() and gauss() functions from the python base random module.
Shuffle the attributes of the input individual and return the mutant. The individual is expected to be a sequence. The indpb argument is the probability of each attribute to be moved. Usually this mutation is applied on vector of indices.
| Parameters: |
|
|---|---|
| Returns: | A tuple of one individual. |
This function uses the random() and randint() functions from the python base random module.
Flip the value of the attributes of the input individual and return the mutant. The individual is expected to be a sequence and the values of the attributes shall stay valid after the not operator is called on them. The indpb argument is the probability of each attribute to be flipped. This mutation is usually applied on boolean individuals.
| Parameters: |
|
|---|---|
| Returns: | A tuple of one individual. |
This function uses the random() function from the python base random module.
Mutate an individual by replacing attributes, with probability indpb, by a integer uniformly drawn between low and up inclusively.
| Parameters: |
|
|---|---|
| Returns: | A tuple of one individual. |
Polynomial mutation as implemented in original NSGA-II algorithm in C by Deb.
| Parameters: |
|
|---|---|
| Returns: | A tuple of one individual. |
Mutate an evolution strategy according to its strategy
attribute as described in [Beyer2002]. First the strategy is mutated
according to an extended log normal rule, ![\boldsymbol{\sigma}_t =
\exp(\tau_0 \mathcal{N}_0(0, 1)) \left[ \sigma_{t-1, 1}\exp(\tau
\mathcal{N}_1(0, 1)), \ldots, \sigma_{t-1, n} \exp(\tau
\mathcal{N}_n(0, 1))\right]](../_images/math/0ec9c2b7621c1044613c554e09c646070850c949.png) , with
, with 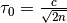 and
and 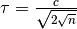 ,
the the individual is mutated by a normal distribution of mean 0 and
standard deviation of
,
the the individual is mutated by a normal distribution of mean 0 and
standard deviation of 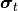 (its current
strategy) then . A recommended choice is c=1 when using a
(its current
strategy) then . A recommended choice is c=1 when using a 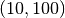 evolution strategy [Beyer2002] [Schwefel1995].
evolution strategy [Beyer2002] [Schwefel1995].
| Parameters: |
|
|---|---|
| Returns: | A tuple of one individual. |
| [Beyer2002] | (1, 2) Beyer and Schwefel, 2002, Evolution strategies - A Comprehensive Introduction |
| [Schwefel1995] | Schwefel, 1995, Evolution and Optimum Seeking. Wiley, New York, NY |
This operator shrinks the individual by chosing randomly a branch and replacing it with one of the branch’s arguments (also randomly chosen).
| Parameters: | individual – The tree to be shrinked. |
|---|---|
| Returns: | A tuple of one tree. |
Randomly select a point in the tree individual, then replace the subtree at that point as a root by the expression generated using method expr().
| Parameters: |
|
|---|---|
| Returns: | A tuple of one tree. |
Replaces a randomly chosen primitive from individual by a randomly chosen primitive with the same number of arguments from the pset attribute of the individual.
| Parameters: | individual – The normal or typed tree to be mutated. |
|---|---|
| Returns: | A tuple of one tree. |
This operator works on the constants of the tree individual. In mode "one", it will change the value of one of the individual ephemeral constants by calling its generator function. In mode "all", it will change the value of all the ephemeral constants.
| Parameters: |
|
|---|---|
| Returns: | A tuple of one tree. |
Inserts a new branch at a random position in individual. The subtree at the chosen position is used as child node of the created subtree, in that way, it is really an insertion rather than a replacement. Note that the original subtree will become one of the children of the new primitive inserted, but not perforce the first (its position is randomly selected if the new primitive has more than one child).
| Parameters: | individual – The normal or typed tree to be mutated. |
|---|---|
| Returns: | A tuple of one tree. |
Select k individuals from the input individuals using k tournaments of tournsize individuals. The list returned contains references to the input individuals.
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
This function uses the choice() function from the python base random module.
Select k individuals from the input individuals using k spins of a roulette. The selection is made by looking only at the first objective of each individual. The list returned contains references to the input individuals.
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
This function uses the random() function from the python base random module.
Warning
The roulette selection by definition cannot be used for minimization or when the fitness can be smaller or equal to 0.
Apply NSGA-II selection operator on the individuals. Usually, the size of individuals will be larger than k because any individual present in individuals will appear in the returned list at most once. Having the size of individuals equals to k will have no effect other than sorting the population according to their front rank. The list returned contains references to the input individuals. For more details on the NSGA-II operator see [Deb2002].
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
| [Deb2002] | Deb, Pratab, Agarwal, and Meyarivan, “A fast elitist non-dominated sorting genetic algorithm for multi-objective optimization: NSGA-II”, 2002. |
Apply SPEA-II selection operator on the individuals. Usually, the size of individuals will be larger than n because any individual present in individuals will appear in the returned list at most once. Having the size of individuals equals to n will have no effect other than sorting the population according to a strength Pareto scheme. The list returned contains references to the input individuals. For more details on the SPEA-II operator see [Zitzler2001].
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
| [Zitzler2001] | Zitzler, Laumanns and Thiele, “SPEA 2: Improving the strength Pareto evolutionary algorithm”, 2001. |
Select k individuals at random from the input individuals with replacement. The list returned contains references to the input individuals.
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
This function uses the choice() function from the python base random module.
Select the k best individuals among the input individuals. The list returned contains references to the input individuals.
| Parameters: |
|
|---|---|
| Returns: | A list containing the k best individuals. |
Select the k worst individuals among the input individuals. The list returned contains references to the input individuals.
| Parameters: |
|
|---|---|
| Returns: | A list containing the k worst individuals. |
Tournament selection which use the size of the individuals in order to discriminate good solutions. This kind of tournament is obviously useless with fixed-length representation, but has been shown to significantly reduce excessive growth of individuals, especially in GP, where it can be used as a bloat control technique (see [Luke2002fighting]). This selection operator implements the double tournament technique presented in this paper.
The core principle is to use a normal tournament selection, but using a special sample function to select aspirants, which is another tournament based on the size of the individuals. To ensure that the selection pressure is not too high, the size of the size tournament (the number of candidates evaluated) can be a real number between 1 and 2. In this case, the smaller individual among two will be selected with a probability size_tourn_size/2. For instance, if size_tourn_size is set to 1.4, then the smaller individual will have a 0.7 probability to be selected.
Note
In GP, it has been shown that this operator produces better results when it is combined with some kind of a depth limit.
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
| [Luke2002fighting] | (1, 2) Luke and Panait, 2002, Fighting bloat with nonparametric parsimony pressure |
Tournament selection based on dominance (D) between two individuals, if the two individuals do not interdominate the selection is made based on crowding distance (CD). The individuals sequence length has to be a multiple of 4. Starting from the beginning of the selected individuals, two consecutive individuals will be different (assuming all individuals in the input list are unique). Each individual from the input list won’t be selected more than twice.
This selection requires the individuals to have a crowding_dist attribute, which can be set by the assignCrowdingDist() function.
| Parameters: |
|
|---|---|
| Returns: | A list of selected individuals. |
Sort the first k individuals into different nondomination levels
using the “Fast Nondominated Sorting Approach” proposed by Deb et al.,
see [Deb2002]. This algorithm has a time complexity of 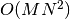 ,
where
,
where  is the number of objectives and
is the number of objectives and  the number of
individuals.
the number of
individuals.
| Parameters: |
|
|---|---|
| Returns: | A list of Pareto fronts (lists), the first list includes nondominated individuals. |
| [Deb2002] | Deb, Pratab, Agarwal, and Meyarivan, “A fast elitist non-dominated sorting genetic algorithm for multi-objective optimization: NSGA-II”, 2002. |
Sort individuals in pareto non-dominated fronts using the Generalized Reduced Run-Time Complexity Non-Dominated Sorting Algorithm presented by Fortin et al. (2013).
| Parameters: | individuals – A list of individuals to select from. |
|---|---|
| Returns: | A list of Pareto fronts (lists), with the first list being the true Pareto front. |
Implement a static limit on some measurement on a GP tree, as defined by Koza in [Koza1989]. It may be used to decorate both crossover and mutation operators. When an invalid (over the limit) child is generated, it is simply replaced by one of its parents, randomly selected.
This operator can be used to avoid memory errors occuring when the tree gets higher than 90 levels (as Python puts a limit on the call stack depth), because it can ensure that no tree higher than this limit will ever be accepted in the population, except if it was generated at initialization time.
| Parameters: |
|
|---|---|
| Returns: | A decorator that can be applied to a GP operator using decorate() |
Note
If you want to reproduce the exact behavior intended by Koza, set key to operator.attrgetter('height') and max_value to 17.
| [Koza1989] | J.R. Koza, Genetic Programming - On the Programming of Computers by Means of Natural Selection (MIT Press, Cambridge, MA, 1992) |
Perform a ring migration between the populations. The migration first select k emigrants from each population using the specified selection operator and then replace k individuals from the associated population in the migarray by the emigrants. If no replacement operator is specified, the immigrants will replace the emigrants of the population, otherwise, the immigrants will replace the individuals selected by the replacement operator. The migration array, if provided, shall contain each population’s index once and only once. If no migration array is provided, it defaults to a serial ring migration (1 – 2 – ... – n – 1). Selection and replacement function are called using the signature selection(populations[i], k) and replacement(populations[i], k). It is important to note that the replacement strategy must select k different individuals. For example, using a traditional tournament for replacement strategy will thus give undesirable effects, two individuals will most likely try to enter the same slot.
| Parameters: |
|
|---|
Object that compiles statistics on a list of arbitrary objects. When created the statistics object receives a key argument that is used to get the values on which the function will be computed. If not provided the key argument defaults to the identity function.
The value returned by the key may be a multi-dimensional object, i.e.: a tuple or a list, as long as the statistical function registered support it. So for example, statistics can be computed directly on multi-objective fitnesses when using numpy statistical function.
| Parameters: | key – A function to access the values on which to compute the statistics, optional. |
|---|
>>> s = Statistics()
>>> s.register("mean", numpy.mean)
>>> s.register("max", max)
>>> s.compile([1, 2, 3, 4])
{'max': 4, 'mean': 2.5}
>>> s.compile([5, 6, 7, 8])
{'max': 8, 'mean': 6.5}
Apply to the input sequence data each registered function and return the results as a dictionnary.
| Parameters: | data – Sequence of objects on which the statistics are computed. |
|---|
Register a function that will be applied on the sequence each time record() is called.
| Parameters: |
|
|---|
Dictionary of Statistics object allowing to compute statistics on multiple keys using a single call to compile(). It takes a set of key-value pairs associating a statistics object to a unique name. This name can then be used to retrieve the statistics object.
The following code computes statistics simultaneously on the length and the first value of the provided objects.
>>> len_stats = Statistics(key=len)
>>> itm0_stats = Statistics(key=itemgetter(0))
>>> mstats = MultiStatistics(length=len_stats, item=itm0_stats)
>>> mstats.register("mean", numpy.mean, axis=0)
>>> mstats.register("max", numpy.max, axis=0)
>>> mstats.compile([[0.0, 1.0, 1.0, 5.0], [2.0, 5.0]])
{'length': {'max': 4, 'mean': 3.0}, 'item': {'max': 2.0, 'mean': 1.0}}
Calls Statistics.compile() with data of each Statistics object.
| Parameters: | data – Sequence of objects on which the statistics are computed. |
|---|
Register a function in each Statistics object.
| Parameters: |
|
|---|
Evolution records as a chronological list of dictionaries.
Data can be retrieved via the select() method given the appropriate names.
The Logbook class may also contain other logbooks refered to as chapters. Chapters are used to store information associated to a specific part of the evolution. For example when computing statistics on different components of individuals (namely MultiStatistics), chapters can be used to distinguish the average fitness and the average size.
Dictionary containing the sub-sections of the logbook which are also Logbook. Chapters are automatically created when the right hand side of a keyworded argument, provided to the record function, is a dictionnary. The keyword determines the chapter’s name. For example, the following line adds a new chapter “size” that will contain the fields “max” and “mean”.
logbook.record(gen=0, size={'max' : 10.0, 'mean' : 7.5})
To access a specific chapter, use the name of the chapter as a dictionnary key. For example, to access the size chapter and select the mean use
logbook.chapters["size"].select("mean")
Compiling a MultiStatistics object returns a dictionary containing dictionnaries, therefore when recording such an object in a logbook using the keyword argument unpacking operator (**), chapters will be automatically added to the logbook.
>>> fit_stats = Statistics(key=attrgetter("fitness.values"))
>>> size_stats = Statistics(key=len)
>>> mstats = MultiStatistics(fitness=fit_stats, size=size_stats)
>>> # [...]
>>> record = mstats.compile(population)
>>> logbook.record(**record)
>>> print logbook
fitness length
------------ ------------
max mean max mean
2 1 4 3
Order of the columns to print when using the stream and __str__() methods. The syntax is a single iterable containing string elements. For example, with the previously defined statistics class, one can print the generation and the fitness average, and maximum with
logbook.header = ("gen", "mean", "max")
If not set the header is built with all fields, in arbritrary order on insertion of the first data. The header can be removed by setting it to None.
Tells the log book to output or not the header when streaming the first line or getting its entire string representation. This defaults True.
Retrieve and delete element index. The header and stream will be adjusted to follow the modification.
| Parameters: | item – The index of the element to remove, optional. It defaults to the first element. |
|---|
You can also use the following syntax to delete elements.
del log[0]
del log[1::5]
Enter a record of event in the logbook as a list of key-value pairs. The informations are appended chronogically to a list as a dictionnary. When the value part of a pair is a dictionnary, the informations contained in the dictionnary are recorded in a chapter entitled as the name of the key part of the pair. Chapters are also Logbook.
Return a list of values associated to the names provided in argument in each dictionary of the Statistics object list. One list per name is returned in order.
>>> log = Logbook()
>>> log.record(gen = 0, mean = 5.4, max = 10.0)
>>> log.record(gen = 1, mean = 9.4, max = 15.0)
>>> log.select("mean")
[5.4, 9.4]
>>> log.select("gen", "max")
([0, 1], [10.0, 15.0])
With a MultiStatistics object, the statistics for each measurement can be retrieved using the chapters member :
>>> log = Logbook()
>>> log.record(**{'gen' : 0, 'fit' : {'mean' : 0.8, 'max' : 1.5},
... 'size' : {'mean' : 25.4, 'max' : 67}})
>>> log.record(**{'gen' : 1, 'fit' : {'mean' : 0.95, 'max' : 1.7},
... 'size' : {'mean' : 28.1, 'max' : 71}})
>>> log.chapters['size'].select("mean")
[25.4, 28.1]
>>> log.chapters['fit'].select("gen", "max")
([0, 1], [1.5, 1.7])
Retrieve the formatted not streamed yet entries of the database including the headers.
>>> log = Logbook()
>>> log.append({'gen' : 0})
>>> print log.stream
gen
0
>>> log.append({'gen' : 1})
>>> print log.stream
1
The hall of fame contains the best individual that ever lived in the population during the evolution. It is lexicographically sorted at all time so that the first element of the hall of fame is the individual that has the best first fitness value ever seen, according to the weights provided to the fitness at creation time.
The insertion is made so that old individuals have priority on new individuals. A single copy of each individual is kept at all time, the equivalence between two individuals is made by the operator passed to the similar argument.
| Parameters: |
|
|---|
The class HallOfFame provides an interface similar to a list (without being one completely). It is possible to retrieve its length, to iterate on it forward and backward and to get an item or a slice from it.
Update the hall of fame with the population by replacing the worst individuals in it by the best individuals present in population (if they are better). The size of the hall of fame is kept constant.
| Parameters: | population – A list of individual with a fitness attribute to update the hall of fame with. |
|---|
Insert a new individual in the hall of fame using the bisect_right() function. The inserted individual is inserted on the right side of an equal individual. Inserting a new individual in the hall of fame also preserve the hall of fame’s order. This method does not check for the size of the hall of fame, in a way that inserting a new individual in a full hall of fame will not remove the worst individual to maintain a constant size.
| Parameters: | item – The individual with a fitness attribute to insert in the hall of fame. |
|---|
Remove the specified index from the hall of fame.
| Parameters: | index – An integer giving which item to remove. |
|---|
Clear the hall of fame.
The Pareto front hall of fame contains all the non-dominated individuals that ever lived in the population. That means that the Pareto front hall of fame can contain an infinity of different individuals.
| Parameters: | similar – A function that tels the Pareto front whether or not two individuals are similar, optional. |
|---|
The size of the front may become very large if it is used for example on a continuous function with a continuous domain. In order to limit the number of individuals, it is possible to specify a similarity function that will return True if the genotype of two individuals are similar. In that case only one of the two individuals will be added to the hall of fame. By default the similarity function is operator.eq().
Since, the Pareto front hall of fame inherits from the HallOfFame, it is sorted lexicographically at every moment.
Update the Pareto front hall of fame with the population by adding the individuals from the population that are not dominated by the hall of fame. If any individual in the hall of fame is dominated it is removed.
| Parameters: | population – A list of individual with a fitness attribute to update the hall of fame with. |
|---|
The History class helps to build a genealogy of all the individuals produced in the evolution. It contains two attributes, the genealogy_tree that is a dictionary of lists indexed by individual, the list contain the indices of the parents. The second attribute genealogy_history contains every individual indexed by their individual number as in the genealogy tree.
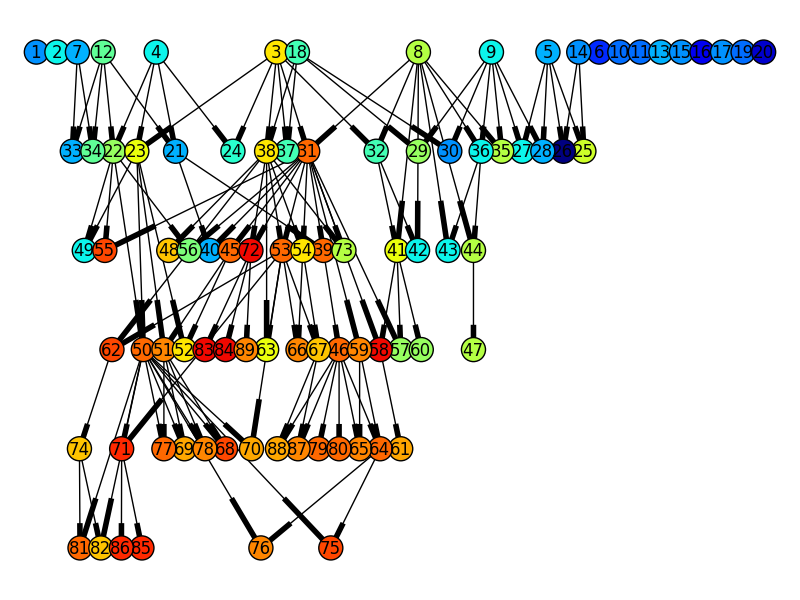
The produced genealogy tree is compatible with NetworkX, here is how to plot the genealogy tree
history = History()
# Decorate the variation operators
toolbox.decorate("mate", history.decorator)
toolbox.decorate("mutate", history.decorator)
# Create the population and populate the history
population = toolbox.population(n=POPSIZE)
history.update(population)
# Do the evolution, the decorators will take care of updating the
# history
# [...]
import matplotlib.pyplot as plt
import networkx
graph = networkx.DiGraph(history.genealogy_tree)
graph = graph.reverse() # Make the grah top-down
colors = [toolbox.evaluate(history.genealogy_history[i])[0] for i in graph]
networkx.draw(graph, node_color=colors)
plt.show()
Using NetworkX in combination with pygraphviz (dot layout) this amazing genealogy tree can be obtained from the OneMax example with a population size of 20 and 5 generations, where the color of the nodes indicate there fitness, blue is low and red is high.
Note
The genealogy tree might get very big if your population and/or the number of generation is large.
Update the history with the new individuals. The index present in their history_index attribute will be used to locate their parents, it is then modified to a unique one to keep track of those new individuals. This method should be called on the individuals after each variation.
| Parameters: | individuals – The list of modified individuals that shall be inserted in the history. |
|---|
If the individuals do not have a history_index attribute, the attribute is added and this individual is considered as having no parent. This method should be called with the initial population to initialize the history.
Modifying the internal genealogy_index of the history or the history_index of an individual may lead to unpredictable results and corruption of the history.
Property that returns an appropriate decorator to enhance the operators of the toolbox. The returned decorator assumes that the individuals are returned by the operator. First the decorator calls the underlying operation and then calls the update() function with what has been returned by the operator. Finally, it returns the individuals with their history parameters modified according to the update function.
Provide the genealogy tree of an individual. The individual must have an attribute history_index as defined by update() in order to retrieve its associated genealogy tree. The returned graph contains the parents up to max_depth variations before this individual. If not provided the maximum depth is up to the begining of the evolution.
| Parameters: |
|
|---|---|
| Returns: | A dictionary where each key is an individual index and the values are a tuple corresponding to the index of the parents. |